CovReport v1

Description
CovReport is a new tool to visualize the coverage data of a diagnostic sequencing test.
It generates a one-glance overview of exon-level coverage that can be directly annexed to a diagnostic report.
The article describing this tool was published in
Scientific Reports
A new tool CovReport generates easy-to-understand sequencing coverage summary for diagnostic reports
Sci Rep 10-Apr-2020; 10(1):6247; DOI: 10.1038/s41598-020-63079-4; PMID: 32277129
This work was also presented
at the European Human Genetics Conference
in Gothenburg, Sweden June 15-18, 2019.
The tool has been implemented as a standalone Java application and can run on any platform where
Java Runtime (JRE) is installed (Windows, Mac or Linux). The supported Java version is 8.
Background
There is a critical need to make diagnostic reports of high-throughput sequencing extremely clear
to avoid any misinterpretation. Target sequence coverage is a key quality control information for
a sequencing test, since important clinical decisions are made based on this data. If coverage
is not sufficient even for a small region of a highly suspected candidate gene, an additional
sequencing test is needed to make sure that the pathogenic variant is not missed.
Download
counter
Click here
to download CovReport application in a compressed format.
Make sure that you agree with Terms and Condition before you start to download or/and use the Software.
The downloaded file CovReport.zip is a compressed file.
This ZIP file should be extracted into local hard drive working folder, for example, C:\CovReport.
The content of this working folder is the following:
msg - folder with internalization message files used for generated PDF file
(see also Language section)CovReport.jar - Java executable archiverun.cmd - Windows command to start the applicationrunFromCommandLine.cmd - Windows command line helper
After the first execution of CovReport application the following items are created in the working folder:
pdf-results - folder with PDF files generated by the applicationCovReport.config - file with configuration persistence data; this file could be manually updated and reused to replace default for command line execution
The application could be started via User Interface (UI) or from command line. The command line options are:
-i,--input <arg> input file
-n,--name <arg> patient name
-c,--config <arg> config file (optional)
|
The optional config file parameter is CovReport.config generated by the application started in UI mode.
Input file format
An example input CSV file could be found here.
Input file is a tab delimited CSV file.
All entries in the file can be quoted per CSV file standard or can be without quotes
(special characters or delimiters are not expected in input file).
The first line containing column headers is ignored.
All other file lines are one line per single exon data.
The following columns are expected in the input file in the specified order:
RefSeqName - string, sequence reference nameGeneSymbol - string, gene symbolExon - positive whole number, exon's numberSize - positive whole number, exon size in base pairs countMean Depth - number with fraction, not usedSD Depth - number with fraction, not usedCoverage 1x - number with fraction in range 0..100
The number could have comma or dot fraction delimiter, e.g. 12.34 or 12,34Coverage 5x - same as aboveCoverage 10x - same as aboveCoverage 20x - same as aboveCoverage 30x - same as above
User Interface (UI)
After launching the application the following minimum steps are required to generate a PDF report:
- Load source exon coverage file. See details about this file format in section Input file format
- Enter patient name
- Generate report
The example generated PDF file could be found here.
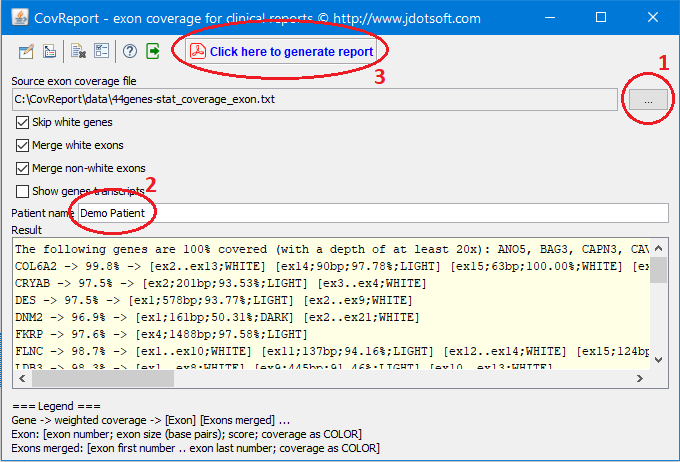
Configuration
The application could be configured with the following options:
- Show report in console
- Choose coverage { 1x, 5x, 10, 20x, 30x }
- Skip white genes
- Merge white exons
- Merge non-white exons
- Show genes transcripts
The following options are for generated PDF file:
- Show input file name { none, simple, absolute file name }
- Draw exon size as scaled or all exons the same size
- Show gene weighted coverage
- Draw merged exons same size
- Show statistics
- Show report date
All configuration options are preserved in CovReport.config file for the next execution.
Configuration options are available in following two places:
- Main window - options which are modified often. See User Interface (UI) section.
- Settings window - options which are modified rarely.
Click the button  in the toolbar to open Settings popup: in the toolbar to open Settings popup:
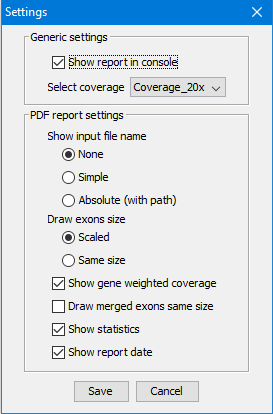
Language
All text in the generated PDF file is in the current locale language.
The following languages are supported in the current distribution: English and French.
The fallback language is English in case of the current locale is not supported.
New locale language can be added by creating file message_NN.properties
in CovReport/msg folder for the NN locale.
Feel free to provide these files to us to be included in future distributions.
The text included in the generated PDF file can be customized by modifying the message file.
Manual override
The exon coverage data loaded from the input file can be overridden with data obtained from additional sequencing experiments
(e.g. Sanger sequencing of poorly covered exons).
Click the button  in the toolbar to manually override: in the toolbar to manually override:
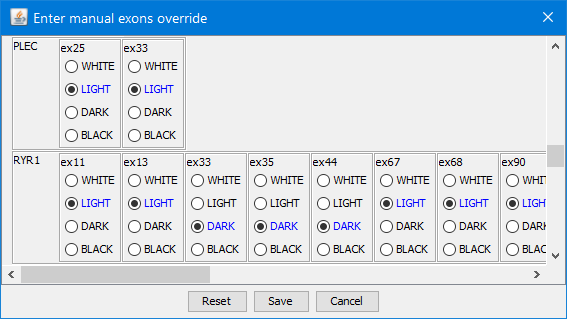
The blue options are default values from the input file.
Any edits could be reset to the default state by pressing button Reset.
The saved edits are preserved in a CSV file with suffix .override next to the input file.
For example, input and override file:
44genes-stat_coverage_exon.txt - input file44genes-stat_coverage_exon.override.txt - override file
The exon with updated coverage will be marked with an asterisk in the final PDF file,
like in the example below for exon 1 which was manually overriden:

Comments
The generated PDF report could be amended with comments.
Click the button  in the toolbar to enter comments: in the toolbar to enter comments:
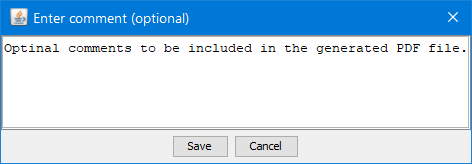
References
The following actions are available in the CovReport application:
 Browse for input Source exon coverage file. This should be the first step to generate PDF report file.
Browse for input Source exon coverage file. This should be the first step to generate PDF report file.
 Add comments to the PDF report file. This icon is disabled if no input file is provided.
See details in Comments section;
Add comments to the PDF report file. This icon is disabled if no input file is provided.
See details in Comments section;
 Manually override input data. This icon is disabled if no input file is provided.
See details in Manual override section;
Manually override input data. This icon is disabled if no input file is provided.
See details in Manual override section;
 Reset application. The input Source exon coverage file and Patient name will be clearted.
All configuration data is remaining intact.
Reset application. The input Source exon coverage file and Patient name will be clearted.
All configuration data is remaining intact.
 Open Settings popup window.
See details in Configuration section;
Open Settings popup window.
See details in Configuration section;
 Open About CovReport popup window with version, build and copyright.
Open About CovReport popup window with version, build and copyright.
 Updates
Updates CovReport.config file with current configuration and closes the application.
 Generates PDF report file.
This button is hidden until Source exon coverage file and Patient name are provided.
Generates PDF report file.
This button is hidden until Source exon coverage file and Patient name are provided.
Open source dependencies
The application is using external open source project libraries (JARs) which are embedded into the main executable JAR:
• Apache PdfBox
• Apache CLI
• JarClassLoader
Troubleshooting
 Application does not start. Application does not start.
• Verify that Java Runtime (JRE) is installed. Check Java verison. It should be 8 or higher.
• Use runDebug.cmd on Windows to check the startup exception displayed in command window.
 Generated PDF report file is not opened automatically. Generated PDF report file is not opened automatically.
• Verify that PDF Acrobat reader is installed.
• In rare cases the default mechanism to auto open the generated PDF file is not working.
This was observed on Mac and Linux.
Try to add the following line into the CovReport.config file:
pdf_auto_open=N
where N is a number from 0 to 4 (0 is a default).
Terms and Conditions of Use
Please find Terms and Conditions of Use document here.
You must accept and follow these Terms and Conditions of Use when using this website
and the CovReport application.
|



